Health datasets play a vital position in analysis and medical training, however it may be difficult to create a dataset that represents the true world. For instance, dermatology circumstances are numerous of their look and severity and manifest otherwise throughout pores and skin tones. Yet, present dermatology picture datasets typically lack illustration of on a regular basis circumstances (like rashes, allergy symptoms and infections) and skew in the direction of lighter pores and skin tones. Furthermore, race and ethnicity data is steadily lacking, hindering our capacity to evaluate disparities or create options.
To handle these limitations, we’re releasing the Skin Condition Image Network (SCIN) dataset in collaboration with physicians at Stanford Medicine. We designed SCIN to mirror the broad vary of considerations that folks search for on-line, supplementing the sorts of circumstances usually present in medical datasets. It incorporates images throughout numerous pores and skin tones and physique components, serving to to make sure that future AI instruments work successfully for all. We’ve made the SCIN dataset freely out there as an open-access resource for researchers, educators, and builders, and have taken cautious steps to guard contributor privateness.
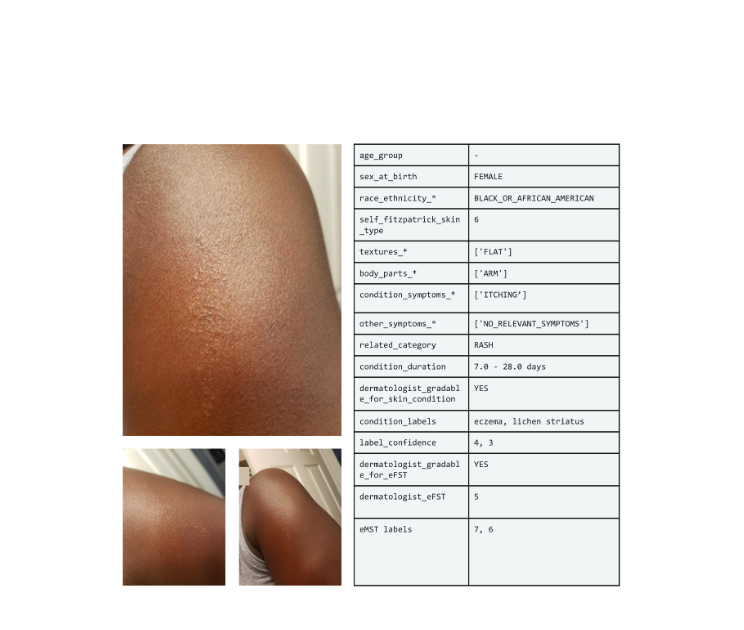
 |
| Example set of images and metadata from the SCIN dataset. |
Dataset composition
The SCIN dataset at the moment incorporates over 10,000 images of pores and skin, nail, or hair circumstances, straight contributed by people experiencing them. All contributions have been made voluntarily with knowledgeable consent by people within the US, below an institutional-review board authorised research. To present context for retrospective dermatologist labeling, contributors have been requested to take images each close-up and from barely additional away. They got the choice to self-report demographic data and tanning propensity (self-reported Fitzpatrick Skin Type, i.e., sFST), and to explain the feel, period and signs associated to their concern.
One to 3 dermatologists labeled every contribution with as much as 5 dermatology circumstances, together with a confidence rating for every label. The SCIN dataset incorporates these particular person labels, in addition to an aggregated and weighted differential prognosis derived from them that may very well be helpful for mannequin testing or coaching. These labels have been assigned retrospectively and are usually not equal to a medical prognosis, however they permit us to match the distribution of dermatology circumstances within the SCIN dataset with present datasets.
 |
| The SCIN dataset incorporates largely allergic, inflammatory and infectious circumstances whereas datasets from medical sources deal with benign and malignant neoplasms. |
While many present dermatology datasets deal with malignant and benign tumors and are meant to help with pores and skin most cancers prognosis, the SCIN dataset consists largely of widespread allergic, inflammatory, and infectious circumstances. The majority of images within the SCIN dataset present early-stage considerations — greater than half arose lower than per week earlier than the picture, and 30% arose lower than a day earlier than the picture was taken. Conditions inside this time window are seldom seen inside the well being system and subsequently are underrepresented in present dermatology datasets.
We additionally obtained dermatologist estimates of Fitzpatrick Skin Type (estimated FST or eFST) and layperson labeler estimates of Monk Skin Tone (eMST) for the images. This allowed comparability of the pores and skin situation and pores and skin kind distributions to these in present dermatology datasets. Although we didn’t selectively goal any pores and skin sorts or pores and skin tones, the SCIN dataset has a balanced Fitzpatrick pores and skin kind distribution (with extra of Types 3, 4, 5, and 6) in comparison with comparable datasets from medical sources.
 |
| Self-reported and dermatologist-estimated Fitzpatrick Skin Type distribution within the SCIN dataset in contrast with present un-enriched dermatology datasets (Fitzpatrick17k, PH², SKINL2, and PAD-UFES-20). |
The Fitzpatrick Skin Type scale was initially developed as a photo-typing scale to measure the response of pores and skin sorts to UV radiation, and it’s broadly utilized in dermatology analysis. The Monk Skin Tone scale is a more moderen 10-shade scale that measures pores and skin tone fairly than pores and skin phototype, capturing extra nuanced variations between the darker pores and skin tones. While neither scale was meant for retrospective estimation utilizing images, the inclusion of those labels is meant to allow future analysis into pores and skin kind and tone illustration in dermatology. For instance, the SCIN dataset offers an preliminary benchmark for the distribution of those pores and skin sorts and tones within the US inhabitants.
The SCIN dataset has a excessive illustration of ladies and youthful people, probably reflecting a mixture of things. These might embody variations in pores and skin situation incidence, propensity to hunt well being data on-line, and variations in willingness to contribute to analysis throughout demographics.
Crowdsourcing technique
To create the SCIN dataset, we used a novel crowdsourcing technique, which we describe within the accompanying analysis paper co-authored with investigators at Stanford Medicine. This method empowers people to play an energetic position in healthcare analysis. It permits us to achieve individuals at earlier levels of their well being considerations, doubtlessly earlier than they search formal care. Crucially, this technique makes use of commercials on internet search consequence pages — the start line for many individuals’s well being journey — to attach with members.
Our outcomes display that crowdsourcing can yield a high-quality dataset with a low spam charge. Over 97.5% of contributions have been real images of pores and skin circumstances. After performing additional filtering steps to exclude images that have been out of scope for the SCIN dataset and to take away duplicates, we have been capable of launch practically 90% of the contributions obtained over the 8-month research interval. Most images have been sharp and well-exposed. Approximately half of the contributions embody self-reported demographics, and 80% include self-reported data regarding the pores and skin situation, corresponding to texture, period, or different signs. We discovered that dermatologists’ capacity to retrospectively assign a differential prognosis depended extra on the supply of self-reported data than on picture high quality.
 |
| Dermatologist confidence of their labels (scale from 1-5) relied on the supply of self-reported demographic and symptom data. |
While excellent picture de-identification can by no means be assured, defending the privateness of people who contributed their images was a high precedence when creating the SCIN dataset. Through knowledgeable consent, contributors have been made conscious of potential re-identification dangers and suggested to keep away from importing images with figuring out options. Post-submission privateness safety measures included handbook redaction or cropping to exclude doubtlessly figuring out areas, reverse picture searches to exclude publicly out there copies and metadata elimination or aggregation. The SCIN Data Use License prohibits makes an attempt to re-identify contributors.
We hope the SCIN dataset will probably be a useful resource for these working to advance inclusive dermatology analysis, training, and AI device growth. By demonstrating an alternative choice to conventional dataset creation strategies, SCIN paves the way in which for extra representative datasets in areas the place self-reported knowledge or retrospective labeling is possible.
Acknowledgements
We are grateful to all our co-authors Abbi Ward, Jimmy Li, Julie Wang, Sriram Lakshminarasimhan, Ashley Carrick, Bilson Campana, Jay Hartford, Pradeep Kumar S, Tiya Tiyasirisokchai, Sunny Virmani, Renee Wong, Yossi Matias, Greg S. Corrado, Dale R. Webster, Dawn Siegel (Stanford Medicine), Steven Lin (Stanford Medicine), Justin Ko (Stanford Medicine), Alan Karthikesalingam and Christopher Semturs. We additionally thank Yetunde Ibitoye, Sami Lachgar, Lisa Lehmann, Javier Perez, Margaret Ann Smith (Stanford Medicine), Rachelle Sico, Amit Talreja, Annisah Um’rani and Wayne Westerlind for their important contributions to this work. Finally, we’re grateful to Heather Cole-Lewis, Naama Hammel, Ivor Horn, Michael Howell, Yun Liu, and Eric Teasley for their insightful feedback on the research design and manuscript.