Despite current progress within the discipline of medical synthetic intelligence (AI), most current fashions are slender, single-task techniques that require massive portions of labeled information to coach. Moreover, these fashions can’t be simply reused in new scientific contexts as they typically require the gathering, de-identification and annotation of site-specific information for each new deployment atmosphere, which is each laborious and costly. This downside of data-efficient generalization (a mannequin’s capacity to generalize to new settings utilizing minimal new information) continues to be a key translational problem for medical machine studying (ML) fashions and has in flip, prevented their broad uptake in actual world healthcare settings.
The emergence of basis fashions affords a big alternative to rethink growth of medical AI to make it extra performant, safer, and equitable. These fashions are skilled utilizing information at scale, typically by self-supervised studying. This course of leads to generalist fashions that may quickly be tailored to new duties and environments with much less want for supervised information. With basis fashions, it could be doable to securely and effectively deploy fashions throughout numerous scientific contexts and environments.
In “Robust and Efficient MEDical Imaging with Self-supervision” (REMEDIS), to be printed in Nature Biomedical Engineering, we introduce a unified large-scale self-supervised studying framework for constructing basis medical imaging fashions. This technique combines massive scale supervised switch studying with self-supervised studying and requires minimal task-specific customization. REMEDIS reveals vital enchancment in data-efficient generalization throughout medical imaging duties and modalities with a 3–100x discount in site-specific information for adapting fashions to new scientific contexts and environments. Building on this, we’re excited to announce Medical AI Research Foundations (hosted by PhysioNet), an growth of the general public launch of chest X-ray Foundations in 2022. Medical AI Research Foundations is a set of open-source non-diagnostic fashions (beginning with REMEDIS fashions), APIs, and assets to assist researchers and builders speed up medical AI analysis.
Large scale self-supervision for medical imaging
REMEDIS makes use of a mix of pure (non-medical) pictures and unlabeled medical pictures to develop sturdy medical imaging basis fashions. Its pre-training technique consists of two steps. The first includes supervised illustration studying on a large-scale dataset of labeled pure pictures (pulled from Imagenet 21k or JFT) utilizing the Big Transfer (BiT) technique.
The second step includes intermediate self-supervised studying, which doesn’t require any labels and as a substitute, trains a mannequin to be taught medical information representations independently of labels. The particular strategy used for pre-training and studying representations is SimCLR. The technique works by maximizing settlement between in another way augmented views of the identical coaching instance by way of a contrastive loss in a hidden layer of a feed-forward neural community with multilayer perceptron (MLP) outputs. However, REMEDIS is equally appropriate with different contrastive self-supervised studying strategies. This coaching technique is relevant for healthcare environments as many hospitals purchase uncooked information (pictures) as a routine observe. While processes must be applied to make this information usable inside fashions (i.e., affected person consent previous to gathering the information, de-identification, and so forth.), the pricey, time-consuming, and tough job of labeling that information may very well be prevented utilizing REMEDIS.
 |
| REMEDIS leverages large-scale supervised studying utilizing pure pictures and self-supervised studying utilizing unlabeled medical information to create sturdy basis fashions for medical imaging. |
Given ML mannequin parameter constraints, it’s important that our proposed strategy works when utilizing each small and massive mannequin structure sizes. To examine this intimately, we thought of two ResNet architectures with generally used depth and width multipliers, ResNet-50 (1×) and ResNet-152 (2×) because the spine encoder networks.
After pre-training, the mannequin was fine-tuned utilizing labeled task-specific medical information and evaluated for in-distribution job efficiency. In addition, to guage the data-efficient generalization, the mannequin was additionally optionally fine-tuned utilizing small quantities of out-of-distribution (OOD) information.
 |
| REMEDIS begins with representations initialized utilizing large-scale pure picture pretraining following the Big Transfer (BiT) technique. We then adapt the mannequin to the medical area utilizing intermediate contrastive self-supervised studying with out utilizing any labeled medical information. Finally, we fine-tune the mannequin to particular downstream medical imaging duties. We consider the ML mannequin each in an in-distribution (ID) setting and in an out-of-distribution (OOD) setting to determine the data-efficient generalization efficiency of the mannequin. |
Evaluation and outcomes
To consider the REMEDIS mannequin’s efficiency, we simulate lifelike situations utilizing retrospective de-identified information throughout a broad vary of medical imaging duties and modalities, together with dermatology, retinal imaging, chest X-ray interpretation, pathology and mammography. We additional introduce the notion of data-efficient generalization, capturing the mannequin’s capacity to generalize to new deployment distributions with a considerably lowered want for knowledgeable annotated information from the brand new scientific setting. In-distribution efficiency is measured as (1) enchancment in zero-shot generalization to OOD settings (assessing efficiency in an OOD analysis set, with zero entry to coaching information from the OOD dataset) and (2) vital discount within the want for annotated information from the OOD settings to succeed in efficiency equal to scientific specialists (or threshold demonstrating scientific utility). REMEDIS reveals considerably improved in-distribution efficiency with as much as 11.5% relative enchancment in diagnostic accuracy over a strongly supervised baseline.
More importantly, our technique results in data-efficient generalization of medical imaging fashions, matching sturdy supervised baselines leading to a 3–100x discount within the want for retraining information. While SimCLR is the first self-supervised studying strategy used within the examine, we additionally present that REMEDIS is appropriate with different approaches, similar to MoCo-V2, RELIC and Barlow Twins. Furthermore, the strategy works throughout mannequin structure sizes.
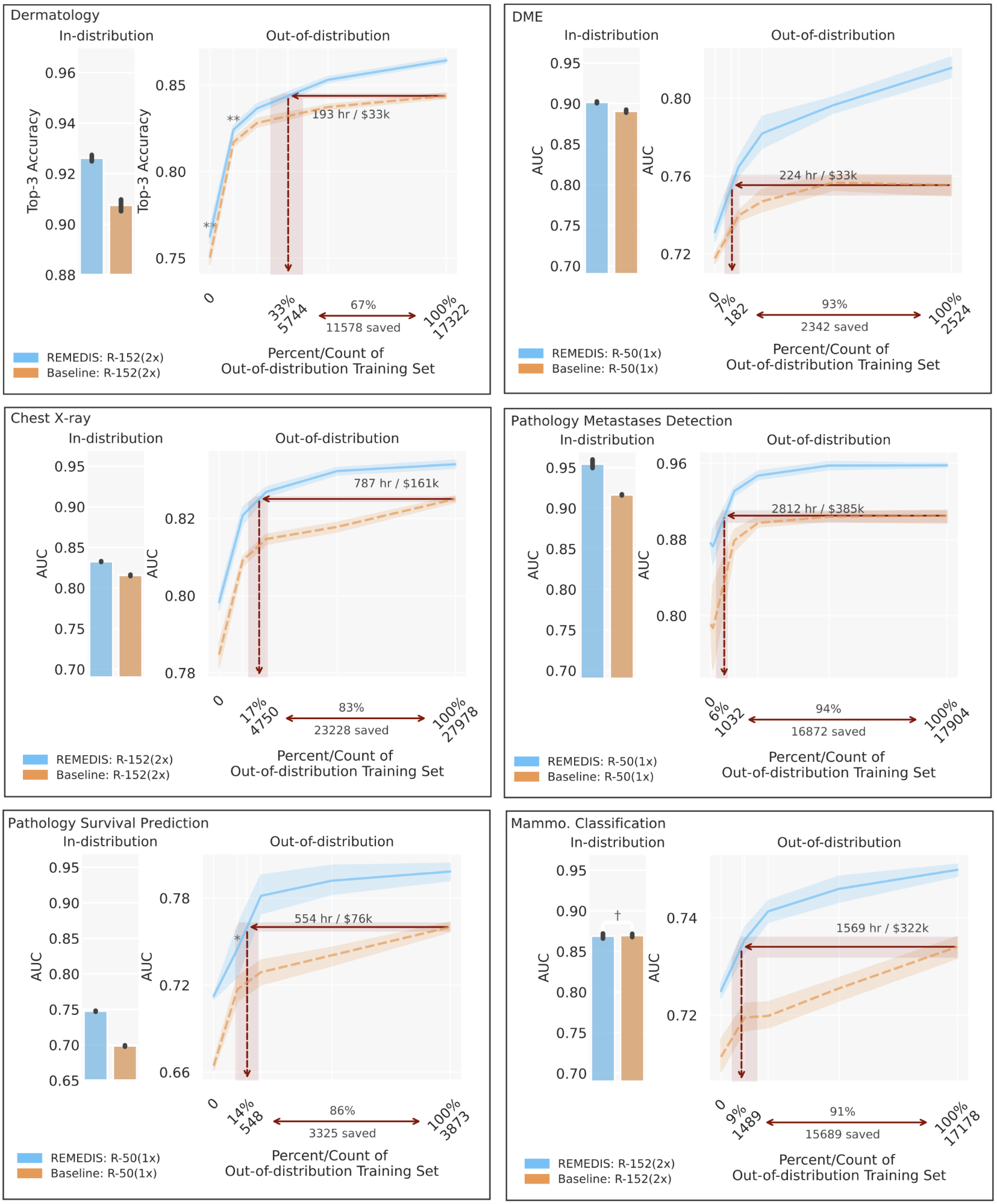 |
| REMEDIS outperformed the supervised baseline pre-trained on JFT-300M for numerous medical duties and demonstrated improved data-efficient generalization, decreasing information wants by 3–100x for adapting fashions to new scientific settings. This might doubtlessly translate to vital discount in clinician hours saved annotating information and value of growing strong medical imaging techniques. |
 |
| REMEDIS is appropriate with MoCo-V2, RELIC and Barlow Twins as alternate self-supervised studying methods. All the REMEDIS variants result in data-efficient generalization enhancements over the sturdy supervised baseline for dermatology situation classification (T1), diabetic macular edema classification (T2), and chest X-ray situation classification (T3). The grey shaded space signifies the efficiency of the sturdy supervised baseline pre-trained on JFT. |
Medical AI Research Foundations
Building on REMEDIS, we’re excited to announce Medical AI Research Foundations, an growth of the general public launch of chest X-ray Foundations in 2022. Medical AI Research Foundations is a repository of open-source medical basis fashions hosted by PhysioNet. This expands the earlier API-based strategy to additionally embody non-diagnostic fashions, to assist researchers and builders speed up their medical AI analysis. We consider that REMEDIS and the discharge of the Medical AI Research Foundations are a step towards constructing medical fashions that may generalize throughout healthcare settings and duties.
We are seeding Medical AI Research Foundations with REMEDIS fashions for chest X-ray and pathology (with associated code). Whereas the prevailing chest X-ray Foundation strategy focuses on offering frozen embeddings for application-specific fantastic tuning from a mannequin skilled on a number of massive personal datasets, the REMEDIS fashions (skilled on public datasets) allow customers to fine-tune end-to-end for his or her software, and to run on native units. We suggest customers check completely different approaches based mostly on their distinctive wants for his or her desired software. We count on so as to add extra fashions and assets for coaching medical basis fashions similar to datasets and benchmarks sooner or later. We additionally welcome the medical AI analysis neighborhood to contribute to this.
Conclusion
These outcomes recommend that REMEDIS has the potential to considerably speed up the event of ML techniques for medical imaging, which might protect their sturdy efficiency when deployed in a wide range of altering contexts. We consider this is a crucial step ahead for medical imaging AI to ship a broad affect. Beyond the experimental outcomes introduced, the strategy and insights described right here have been built-in into a number of of Google’s medical imaging analysis initiatives, similar to dermatology, mammography and radiology amongst others. We’re utilizing an analogous self-supervised studying strategy with our non-imaging basis mannequin efforts, similar to Med-PaLM and Med-PaLM 2.
With REMEDIS, we demonstrated the potential of basis fashions for medical imaging functions. Such fashions maintain thrilling prospects in medical functions with the chance of multimodal illustration studying. The observe of drugs is inherently multimodal and incorporates data from pictures, digital well being information, sensors, wearables, genomics and extra. We consider ML techniques that leverage these information at scale utilizing self-supervised studying with cautious consideration of privateness, security, equity and ethics will assist lay the groundwork for the subsequent technology of studying well being techniques that scale world-class healthcare to everybody.
Acknowledgements
This work concerned in depth collaborative efforts from a multidisciplinary staff of researchers, software program engineers, clinicians, and cross-functional contributors throughout Google Health AI and Google Brain. In specific, we want to thank our first co-author Jan Freyberg and our lead senior authors of those initiatives, Vivek Natarajan, Alan Karthikesalingam, Mohammad Norouzi and Neil Houlsby for his or her invaluable contributions and help. We additionally thank Lauren Winer, Sami Lachgar, Yun Liu and Karan Singhal for his or her suggestions on this publish and Tom Small for help in creating the visuals. Finally, we additionally thank the PhysioNet staff for his or her help on internet hosting Medical AI Research Foundations. Users with questions can attain out to medical-ai-research-foundations at google.com.

