For many years, researchers labored collectively to assemble a whole copy of the molecular directions for a human — a map of the human genome. The first draft was completed in 2000, however with a number of lacking items. Even when a whole reference genome was achieved in 2022, their work was not completed. A single reference genome can’t incorporate recognized genetic variations, comparable to the variants for the gene figuring out whether or not an individual has a blood kind A, B, AB or O. Furthermore, the reference genome didn’t signify the huge range of human ancestries, making it much less helpful for detecting illness or discovering cures for folks from some backgrounds than others. For the previous three years, now we have been half of a world collaboration with 119 scientists throughout 60 establishments, known as the Human Pangenome Research Consortium, to deal with these challenges by creating a brand new and extra consultant map of the human genome, a pangenome.
We are excited to share that right this moment, in “A draft human pangenome reference”, revealed in Nature, this group is saying the completion of the first human pangenome reference. The pangenome combines 47 particular person genome reference sequences and better represents the genomic range of world populations. Building on Google’s deep studying applied sciences and previous advances in genomics, we used instruments primarily based on convolutional neural networks (CNNs) and transformers to sort out the challenges of constructing correct pangenome sequences and utilizing them for genome evaluation. These contributions helped the consortium construct an information-rich useful resource for geneticists, researchers and clinicians round the world.
 |
Using graphs to construct pangenomes
In the typical evaluation workflow for high-throughput DNA sequencing, a sequencing instrument reads thousands and thousands of quick items of a person’s genome, and a program known as a mapper or aligner then estimates the place these items greatest match relative to the single, linear human reference sequence. Next, variant caller software program identifies the distinctive elements of the particular person’s sequence relative to the reference.
But as a result of people carry a various set of sequences, sections which can be current in a person’s DNA however are usually not in the reference genome can’t be analyzed. One examine of 910 African people discovered {that a} whole of 300 million DNA base pairs — 10% of the roughly three billion base pair reference genome — are usually not current in the earlier linear reference however happen in not less than one of the 910 people.
To deal with this problem, the consortium used graph information constructions, that are highly effective for genomics as a result of they’ll signify the sequences of many individuals concurrently, which is required to create a pangenome. Nodes in a graph genome include the recognized set of sequences in a inhabitants, and paths via these nodes compactly describe the distinctive sequences of a person’s DNA.
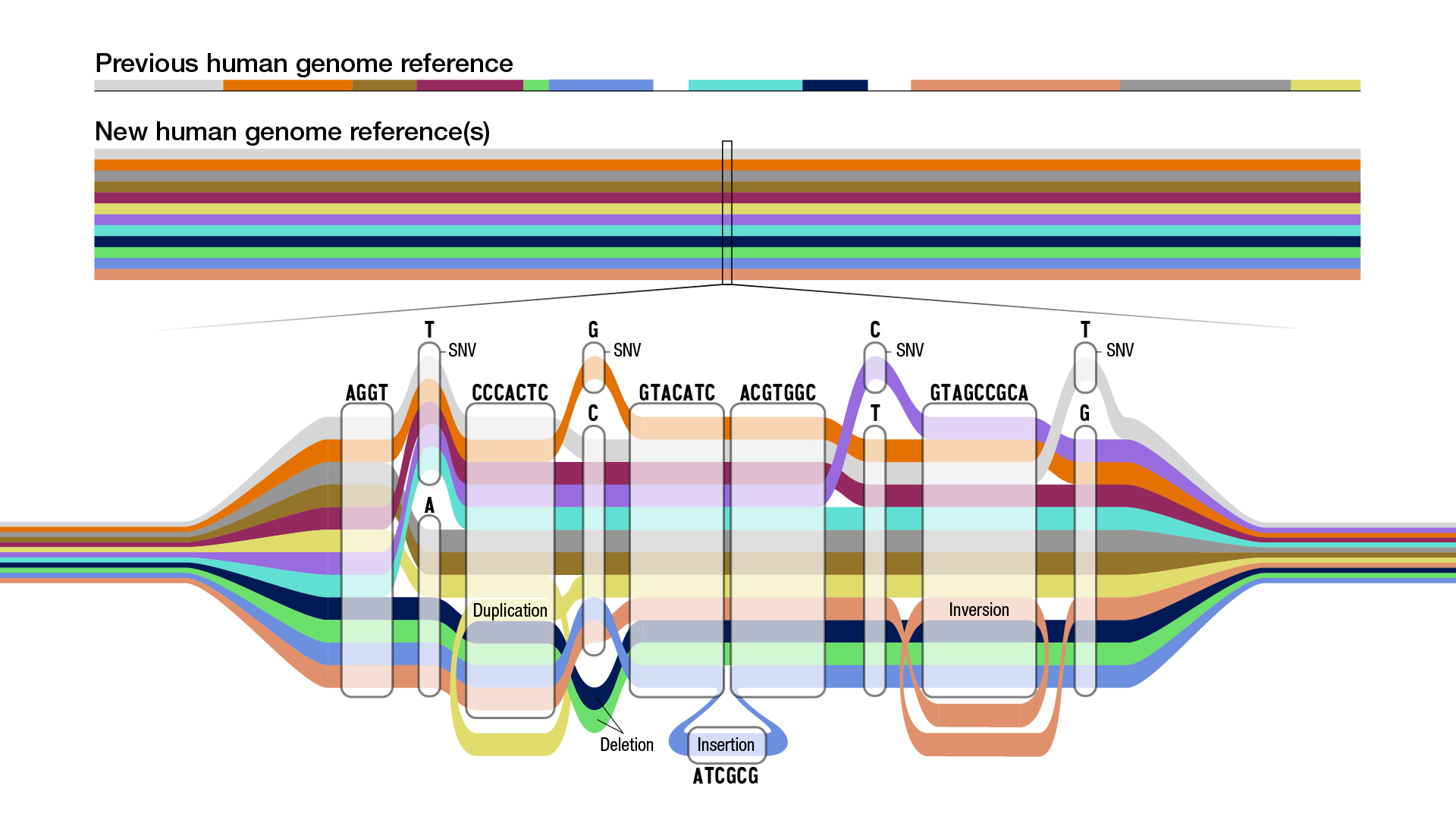 |
| Schematic of a graph genome. Each coloration represents the sequence path of a unique particular person. Multiple paths passing via the identical node point out a number of people share that sequence, however some paths additionally present a single nucleotide variant (SNV), insertions, or deletions. Illustration credit score Darryl Leja, National Human Genome Research Institute (NHGRI). |
 |
| Actual graph genome for the main histocompatibility advanced (MHC) area of the genome. Genes in MHC areas are important to immune perform and are related to an individual’s resistance and susceptibility to infectious illness and autoimmune issues (e.g., ankylosing spondylitis and lupus). The graph reveals the linear human genome reference (inexperienced) and totally different particular person particular person’s sequence (grey). |
Using graphs creates quite a few challenges. They require reference sequences to be extremely correct and the growth of new strategies that may use their information construction as an enter. However, new sequencing applied sciences (comparable to consensus sequencing and phased meeting strategies) have pushed thrilling progress in the direction of fixing these issues.
Long-read sequencing expertise, which reads bigger items of the genome (10,000 to thousands and thousands of DNA characters lengthy) at a time, are important to the creation of top quality reference sequences as a result of bigger items could be stitched collectively into assembled genomes extra simply than the quick items learn out by earlier applied sciences. Short learn sequencing reads items of the genome which can be solely 100 to 300 DNA characters lengthy, however has been the extremely scalable foundation for high-throughput sequencing strategies developed in the 2000s. Though long-read sequencing is newer and has benefits for reference genome creation, many informatics strategies for brief reads hadn’t been developed for lengthy learn applied sciences.
Evolving DeepVariant for error correction
Google initially developed DeepVariant, an open-source CNN variant caller framework that analyzes the short-read sequencing proof of native areas of the genome. However, we had been ready to re-train DeepVariant to yield correct evaluation of Pacific Bioscience’s long-read information.
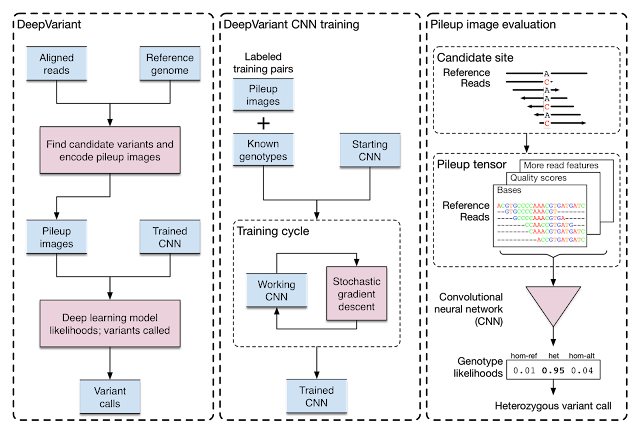 |
| Training and analysis schematic for DeepVariant. |
We subsequent teamed up with researchers at the University of California, Santa Cruz (UCSC) Genomics Institute to take part in a United States Food and Drug Administration competitors for one more long-read sequencing expertise from Oxford Nanopore. Together, we gained the award for highest accuracy in the nanopore class, with a single nucleotide variants (SNVs) accuracy that matched short-read sequencing. This work has been used to detect and deal with genetic illnesses in critically in poor health newborns. The use of DeepVariant on long-read applied sciences supplied the basis for the consortium’s use of DeepVariant for error correction of pangenomes.
DeepVariant’s skill to use a number of long-read sequencing modalities proved helpful for error correction in the Telomere-to-Telomere (T2T) Consortium’s effort that generated the first full meeting of a human genome. Completing this primary genome set the stage to construct the a number of reference genomes required for pangenomes, and T2T was already working intently with the Human Pangenome Project (with many shared members) to scale these practices.
With a set of high-quality human reference genomes on the horizon, growing strategies that would use these assemblies grew in significance. We labored to adapt DeepVariant to use the pangenome developed by the consortium. In partnership with UCSC, we constructed an end-to-end evaluation workflow for graph-based variant detection, and demonstrated improved accuracy throughout a number of thousand samples. The use of the pangenome permits many beforehand missed variants to be appropriately recognized.
 |
| Visualization of variant calls in the KCNE1 gene (a gene with variants related to cardiac arrhythmias and sudden loss of life) utilizing a pangenome reference versus the prior linear reference. Each dot represents a variant name that’s both appropriate (blue dot), incorrect (inexperienced dot) — when a variant is recognized however will not be actually there —or a missed variant name (crimson dot). The prime field reveals variant calls made by DeepVariant utilizing the pangenome reference whereas the backside reveals variant calls made by utilizing the linear reference. Figure tailored from A Draft Human Pangenome Reference. |
Improving pangenome sequences utilizing transformers
Just as new sequencing applied sciences enabled new pangenome approaches, new informatics applied sciences enabled enhancements for sequencing strategies. Google tailored transformer architectures from evaluation of human language to genome sequences to develop DeepConsensus. A key enabler for this was the growth of a differentiable loss perform that would deal with the insertions and deletions widespread in sequencing information. This enabled us to have excessive accuracy with no need a decoder, permitting the pace required to sustain with terabytes of sequencer output.
 |
| Transformer structure for DeepConsensus. DeepConsensus takes as enter the repeated sequence of the DNA molecule, measured from fluorescent gentle detected by the addition of every base. DeepConsensus additionally makes use of as enter the extra detailed details about the sequencing course of, together with the length of the gentle pulse (referred to right here as pulse width or PW), the time between pulses (IP) the signal-to-noise ratio (SN) and which facet of the double helix is being measured (strand). |
 |
| Effect of alignment loss perform in coaching analysis of mannequin output. Better accounting of insertions and deletions by a differentiable alignment perform allows the mannequin coaching course of to better estimate errors. |
DeepConsensus improves the yield and accuracy of instrument information. Because PacBio sequencing gives the main sequence info for the 47 genome assemblies, we might apply DeepConsensus to improve these assemblies. With software of DeepConsensus, consortium members constructed a genome assembler that was ready to attain 99.9997% meeting base-level accuracies.
Conclusion
We developed a number of new approaches to improve genetic sequencing strategies, which we then used to assemble pangenome references that allow extra strong genome evaluation.
But that is simply the starting of the story. In the subsequent stage, a bigger, worldwide group of scientists and clinicians will use this pangenome reference to examine genetic illnesses and make new medicine. And future pangenomes will signify much more people, realizing a imaginative and prescient summarized this fashion in a current Nature story: “Every base, everywhere, all at once.” Read our submit on the Keyword Blog to study extra about the human pangenome reference announcement.
Acknowledgements
Many folks had been concerned in creating the pangenome reference, together with 119 authors throughout 60 organizations, with the Human Pangenome Reference Consortium. This weblog submit highlights Google’s contributions to the broader work. We thank the analysis teams at UCSC Genomics Institute (GI) underneath Professors Benedict Paten and Karen Miga, genome sprucing efforts of Arang Rhie at National Institute of Health (NIH), Genome Assembly and Polishing of Adam Phillipy’s group, and the requirements group at National Institute of Standards and Technology (NIST) of Justin Zook. We thank Google contributors: Pi-Chuan Chang, Maria Nattestad, Daniel Cook, Alexey Kolesnikov, Anastaysia Belyaeva, and Gunjan Baid. We thank John Guilyard for his illustrative animation, and Lizzie Dorfman, Elise Kleeman, Erika Hayden, Cory McLean, Shravya Shetty, Greg Corrado, Katherine Chou, and Yossi Matias for his or her assist, coordination, and management. Last however not least, thanks to the analysis individuals that supplied their DNA to assist construct the pangenome useful resource.

